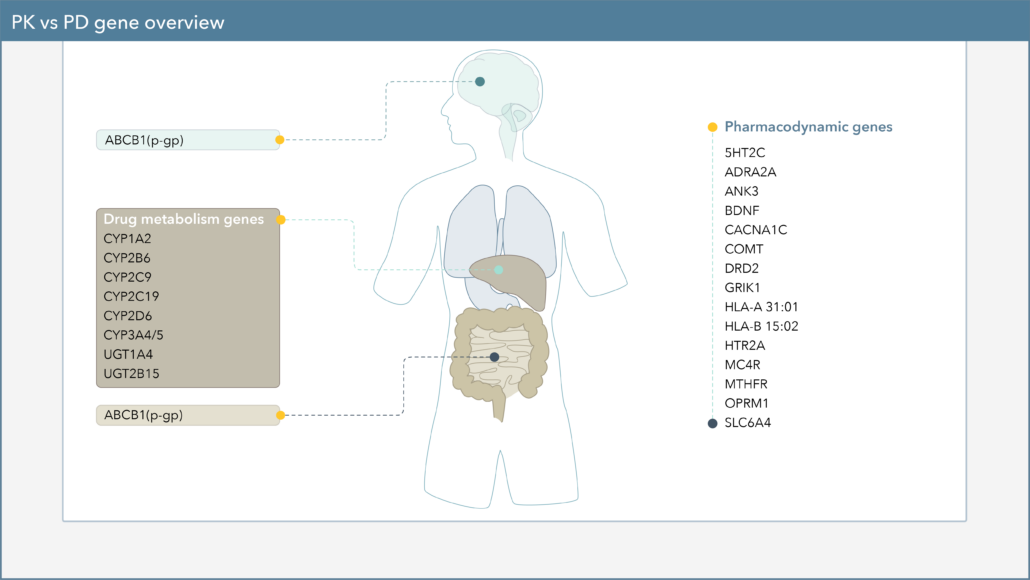
Differences seen in drug response between individuals involves a multitude of factors. One factor that could affect drug response is genetics, particularly variants in pharmacokinetics (PK) and pharmacodynamics (PD) genes.
What are the differences between these two genes?
- Pharmacodynamic genes (pharma=drug + dynamic=how it works)
- Genes that affect therapeutic response or adverse effects to medications
- These genes often express receptors or other proteins that are the molecular targets of these medications.
- Pharmacokinetic genes (pharma=drug + kinetic=how it moves through the body)
-
- Genes which influence how much of a drug is available in the body or brain due to metabolism or absorption
- You can refer back to the article Intro to Pharmacokinetics & CYP Genes to explore this topic in more detail.
Importance of pharmacodynamics and how PD gene variation affects drug response
Pharmacodynamics, sometimes described as “what a drug does to the body”, is the study of the biochemical, physiologic, and molecular effects of drugs on the body and involves receptor binding (including sensitivity) and chemical interactions.1
One way a drug achieves a therapeutic effect is by binding to a receptor, or its target site. Therefore, changes in the sensitivity to receptor binding or receptor expression can influence a drug’s effect.2 Genetic variations in PD genes could result in these changes and thus affect tolerability and/or likelihood of response to drugs.1
Let’s discuss a few examples of PD gene variations that could affect drug response and/or tolerability.
How serotonin transporter gene (SLC6A4) variant affects SSRI drug response
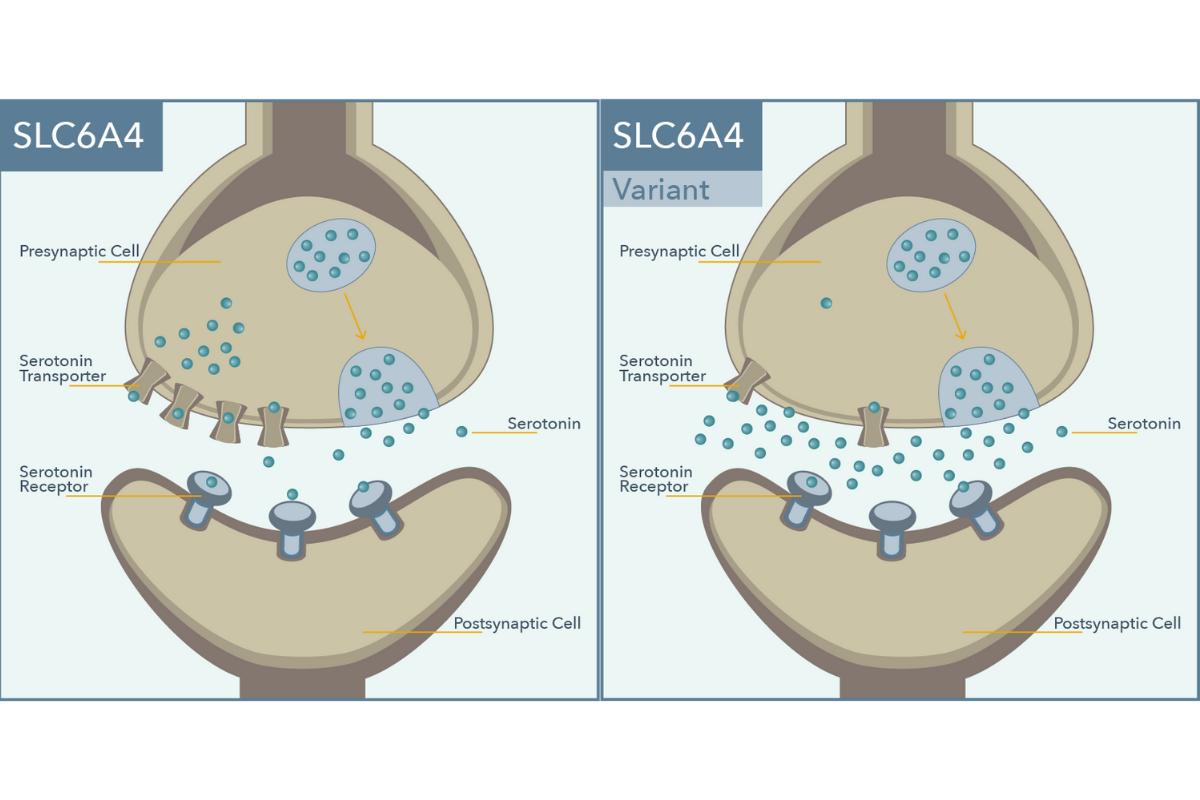
- Physiological role: The serotonin transporter gene, abbreviated SLC6A4, encodes for the protein responsible for the reuptake of serotonin from synapses, the space between neurons.
- Significance: Antidepressant activity of selective serotonin reuptake inhibitor (SSRI) medications is achieved through inhibition of the serotonin transporter. Variation in this gene leads to changes in the amount of serotonin transporters. In the example variant above, serotonin transporter expression is reduced, which is associated with a reduced likelihood of response to, and increased risk of side effects, with SSRIs for depression.3
How μ-opioid receptor gene (OPRM1) variant affects sensitivity to opioids
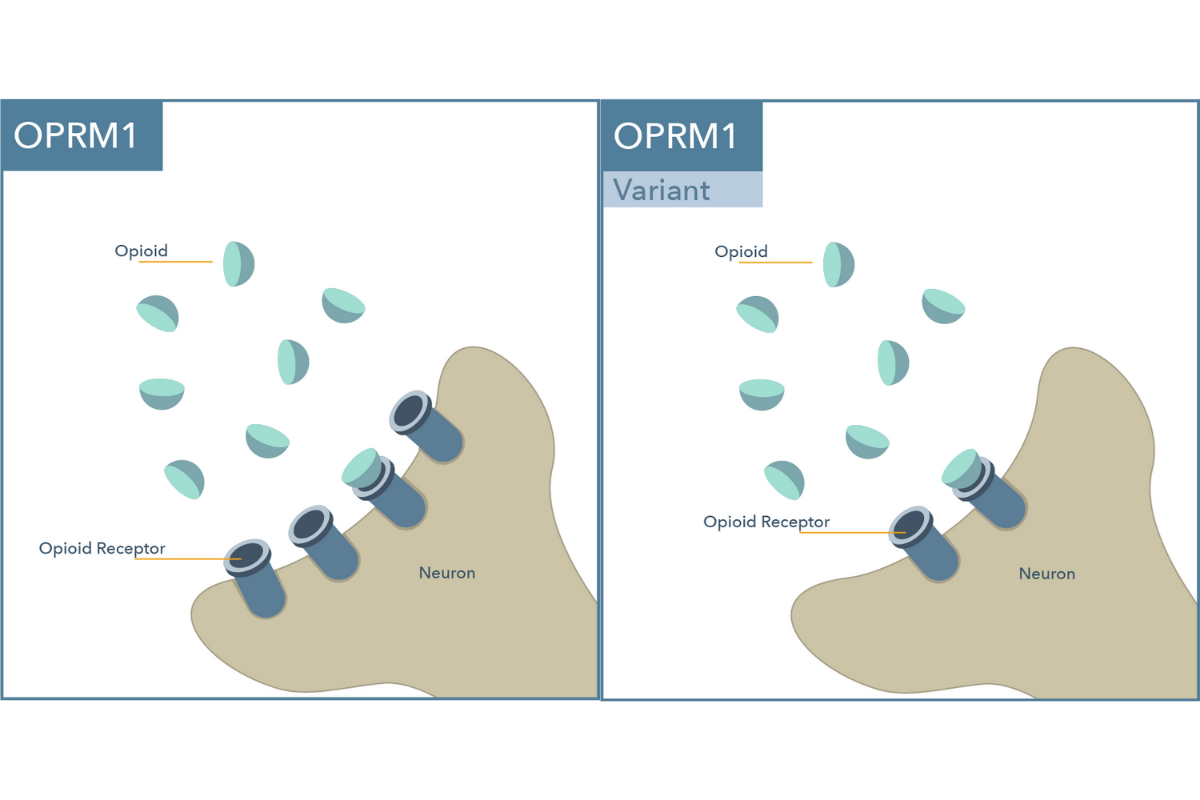
- Physiological role: The μ-opioid receptor gene, abbreviated OPRM1, encodes for a protein that acts as a receptor for opioids in the brain.
- Significance: Variation in this gene affects the expression of OPRM1 receptors and appears to result in different sensitivities to opioids. In the example variant above, OPRM1 receptor expression is reduced, which is associated with decreased sensitivity to opioids and higher dose requirements of opioids for analgesia.4
How melanocortin 4 receptor gene (MC4R) variant can increase weight gain with atypical antipsychotics
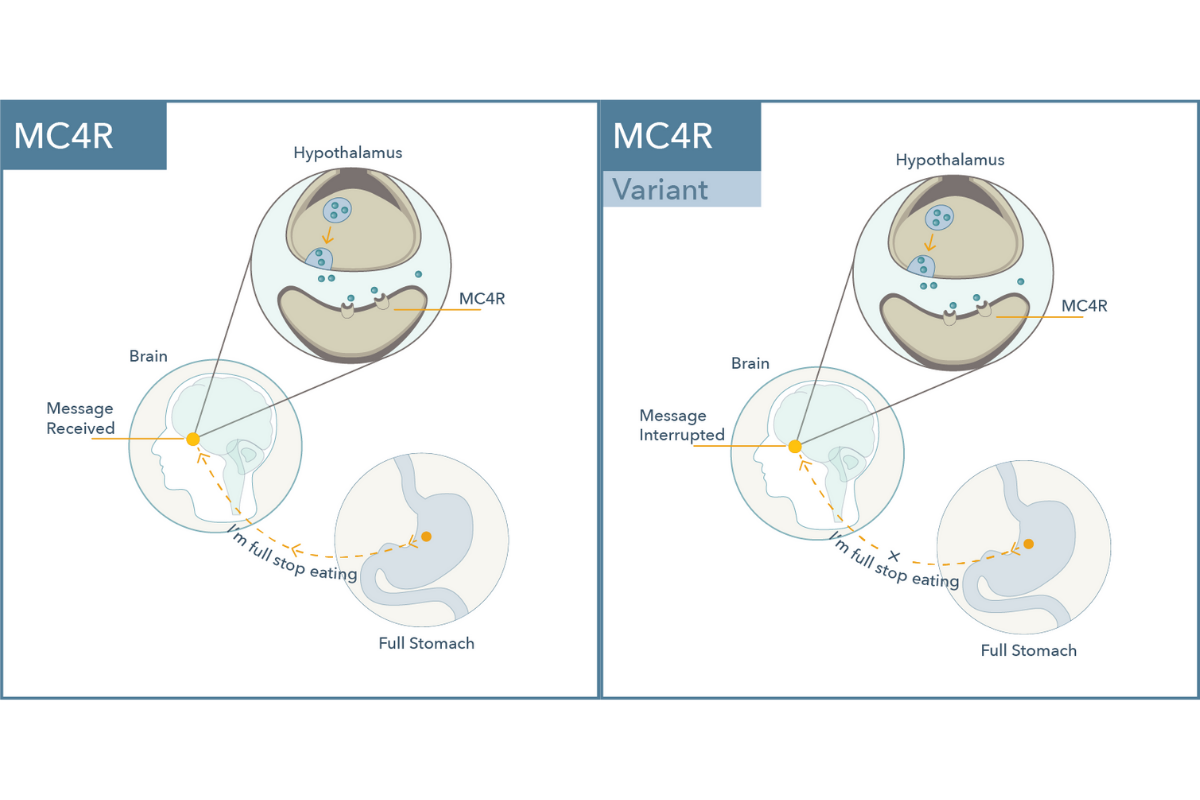
- Physiological role: The melanocortin 4 receptor gene, abbreviated MC4R, encodes for a protein involved in satiety signaling, or control of food intake and energy expenditure.
- Significance: A variant associated with this gene increases the likelihood of gaining weight with the atypical antipsychotics through disruption in satiety signaling.5
In conclusion
These are just a few of the many pharmacodynamic genes that can influence drug response and tolerability in psychiatry. Having insight into an individual’s PD genes can offer a more personalized approach that takes into account both the clinical factors and the patient’s genetics.
For example, SSRIs are commonly used first-line to treat depression, although guidelines such as the American Psychiatric Association Guidelines for Major Depressive Disorder, also include other antidepressants as first-line options (SNRI, mirtazapine, or bupropion).6
It is not uncommon for a clinician to recommend multiple trials of SSRIs before moving on to another antidepressant class. Individuals who have the S/S genotype for SLC6A4 have a lower odds of responding to SSRIs for depression and a greater risk of side effects. Therefore, knowing this information can help maximize likelihood of response and minimize side effects for these individuals by considering other non-SSRI options earlier in the treatment of depression.
Deliver targeted and personalized care with Genomind.
Register with Genomind to use our precision tools and services and help your patients get better. Get started today.
References
- Karczewski, KJ, Daneshjou R, Altman RB. Chapter 7: Pharmacogenomics. PLoS Comput Biol. 2012;8(12):e1002817.
- Merck Manuals. Overview of Pharmacodynamics [Internet] Kenilworth, NJ: Merck Manuals; 2021 [cited 19 Feb 2021]. < https://www.merckmanuals.com/professional/clinical-pharmacology/pharmacodynamics/overview-of-pharmacodynamics#>.
- Porcelli S, Fabbri C, Serretti A. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with antidepressant efficacy. Eur Neuropsychopharmacol. 2012. 22(4):239-58.
- Vieira CM, Fragoso RM, Pereira D, Medeiros R. Pain polymorphisms and opioids: an evidence based review. Mol Med Rep. 2019;19(3):1423-1434.
- Malhotra AK, Correll CU, Chowdhury NI, Müller DJ, Gregersen PK, et al. Association between common variants near the melanocortin 4 receptor gene and severe antipsychotic drug-induced weight gain. Arch Gen Psychiatry. 2012;69(9):904-12.
- Gelenberg AJ, Freeman MP, Markowitz JC, et al. American Psychiatric Association Practice Guideline for the Treatment of Patients With Major Depressive Disorder, Third Edition. Am J Psychiatry. 2010;167(suppl):1-152.