When choosing a medication to treat your patient’s attention-deficit/hyperactivity disorder (ADHD), are you aware that there are several genetic biomarkers that can influence how an individual may respond to and/or tolerate that drug? On the Genomind PGx test you’ll find ADRA2A and COMT, two pharmacodynamic (PD) genes that can impact various pharmacologic treatment options for ADHD.
How pharmacodynamic gene variation affects drug response
Pharmacodynamics, often described as “what a drug does to the body”, is the study of the biochemical, physiologic, and molecular effects of drugs on the body.8 PD gene variability could result in changes to these processes and thus affect medication response and/or tolerability. Let’s discuss two PD genes that have been associated with dopaminergic stimulant response.
What is the alpha-2A adrenergic receptor (ADRA2A)?
The alpha-2A adrenergic receptor (ADRA2A) is concentrated in the prefrontal cortex of the brain and is stimulated by norepinephrine (NE) as illustrated in Figure 1 NE signaling at this receptor is essential for working memory and executive function. Deficits in prefrontal cortex functioning can lead to poor impulse control, distractibility, hyperactivity, forgetfulness, and poor organization and planning.1
Figure 1
How the ADRA2A gene affects methylphenidate response
The therapeutic benefit of methylphenidate is partly mediated by reuptake inhibition of the NE transporter and downstream effects on ADRA2A in the prefrontal cortex. A single nucleotide polymorphism (SNP) at 1291 C > G within the ADRA2A gene has been associated with methylphenidate treatment outcomes in children and adolescents with ADHD.2
The G allele of this SNP in ADRA2A has been associated with an improved response to methylphenidate, particularly for inattentive symptoms in children and adolescents (6-18 years old) with ADHD-I.2 The exact mechanism by which this SNP is connected to methylphenidate response is not fully understood at this time.
A naturalistic PGx study found that following treatment with methylphenidate, carriers of the G allele demonstrated greater improvements in the SNAP-IV inattentive score from baseline at both one month and three months (see Figure 2).4 A meta-analysis conducted by Myer and colleagues revealed that in 355 children and adolescents with ADHD, G allele carriers were 69% more likely to have a response to methylphenidate for ADHD compared to those with the C/C genotype.3 Additionally, the meta-analysis by Hain et al. found that children with ADHD who carry the G allele of the ADRA2A gene showed significantly greater improvements in ADHD symptom scores, specifically on the ADHD Rating Scale-IV (ARS-IV) and the Swanson, Nolan, and Pelham Version-IV Scale (SNAP-IV).
Figure 2
Therapeutic implications of the ADRA2A gene for ADHD treatment
If an individual is an ADRA2A G allele carrier, the Genomind PGx test report will highlight that this variant is associated with an improved response to stimulants (mostly methylphenidate studies) for symptoms of ADHD in children and adolescents as compared to those with the C/C genotype. If clinically indicated, methylphenidate may be considered for ADHD.
If an individual has the C/C genotype (wild-type), the Genomind PGx test report will highlight that this genotype is associated with a reduced response to methylphenidate for symptoms of ADHD in children and adolescents. If clinically appropriate, alternatives to methylphenidate may be considered.
What is catechol-O-methyltransferase (COMT)?
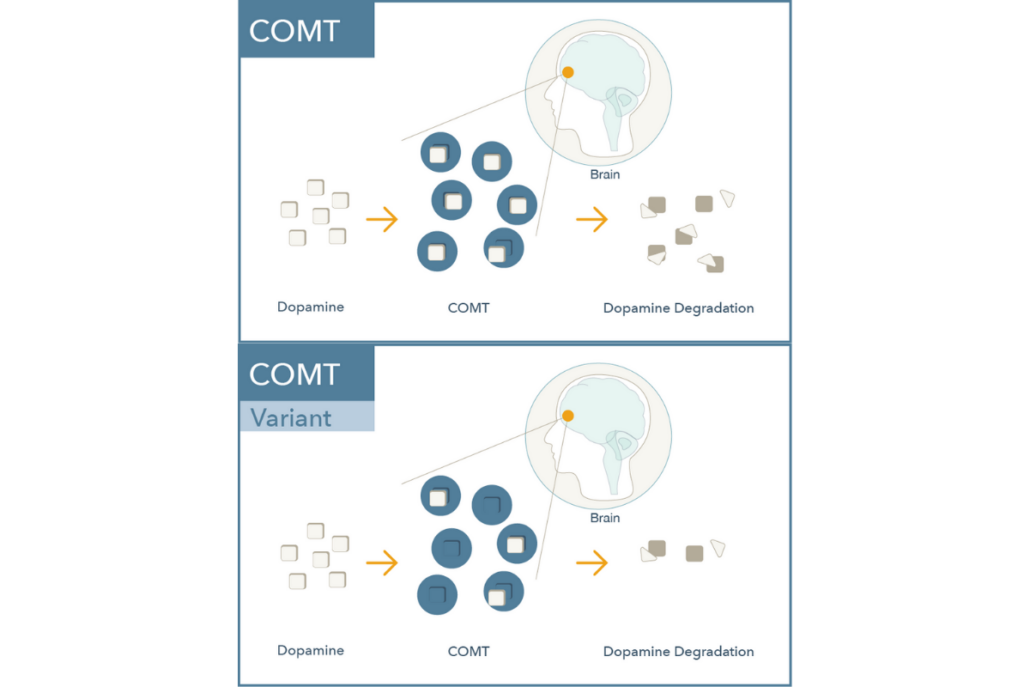
Catechol-O-Methyltransferase (COMT) is a central enzyme that breaks down catecholamines including dopamine, norepinephrine, and epinephrine (see Figure 3). COMT plays a vital role in regulating dopamine signaling in the prefrontal cortex where dopamine transporters (commonly referred to as DAT) are relatively sparse. Dopamine levels here are critical for attention, memory, and other executive functions.5-7
Figure 3
How does the COMT gene affect dopamine?
A SNP in the COMT gene (rs4680, commonly referred to as Val158Met) results in a change of the 158th amino acid in the protein from a valine (Val) to a methionine (Met). This variation is associated with altered enzymatic activity and dopamine levels in the prefrontal cortex.9 As shown below in Figure 4, there are three possible COMT genotypes:
Figure 4
- The Val/Val genotype [high activity] leads to increased COMT enzyme activity and a parallel decrease in frontal cortex dopamine.
- The Val/Met (heterozygous) genotype [normal activity] is considered baseline or a non-clinically actionable genotype.
- The Met/Met genotype [low activity] leads to decreased COMT enzyme activity and a parallel increase in frontal cortex dopamine.
How the COMT gene affects dopaminergic stimulants
If clinically indicated, dopaminergic stimulants (such as methylphenidate or amphetamine) may lead to greater improvements in executive function in individuals with the Val/Val genotype. A meta-analysis revealed that in 599 children and adolescents with ADHD, individuals with the Val/Val genotype were 40% more likely to respond to methylphenidate compared to the Met allele carriers.3
Therapeutic implications of the COMT gene for ADHD treatment
The Genomind PGx test report will highlight the therapeutic implications of an individual’s COMT genotype as follows:
- If an individual has the Val/Val genotype [high activity], dopaminergic stimulants may lead to greater improvements in executive function as compared to Val/Met or Met/Met genotypes. If clinically indicated, dopamine enhancing agents may be considered.
- If an individual has the Val/Met genotype [normal activity], the report will indicate that this genotype confers normal activity and has no known significant clinical impact.
- If an individual has the Met/Met genotype [low activity], the individual may derive less executive function benefit from dopaminergic stimulants as compared to the other genotypes. If clinically appropriate, clinicians may want to assess alternatives to dopaminergic stimulants or augmentation agents.
In conclusion
The Genomind PGx test analyzes important pharmacokinetic and pharmacodynamic genes relevant for the treatment of ADHD, such as ADRA2A and COMT. Specific variants in these pharmacodynamic genes have been associated with the response to dopaminergic stimulants and should be considered in combination with the pharmacokinetic genes that may impact pharmacologic treatments for ADHD. Having insight into an individual’s pharmacogenetics can offer a more personalized approach that takes into account both clinical factors and the patient’s unique genetic predispositions.
Are You Ready to Upgrade Your Practice with Genomind?
Genomind’s pharmacogenetic testing is the most advanced and comprehensive mental health pharmacogenetic test available. Get access to 26 genes related to mental health, 130+ medications, 10+ conditions, state-of-the-art tools, and 360 degrees of support. Register today.
References
- Bidwell CL, Dew RE, Kollins SH. Alpha-2 Adrenergic Receptors and Attention-Deficit/Hyperactivity Disorder. Curr Psychiatry Rep. 2010; 12:366-373
- Polanczyk G, Zeni C, Genro JP, et al. Association of the Adrenergic a2A Receptor Gene with Methylphenidate Improvement of Inattentive Symptoms in Children and Adolescents with Attention-Deficit/Hyperactivity Disorder. Arch Gen Psychiatry. 2007; 64 (2):218-224
- Myer, N. M., Boland, J. R., & Faraone, S. V. (2018). Pharmacogenetics predictors of methylphenidate efficacy in childhood ADHD. Molecular psychiatry, 23(9), 1929–1936.
- Da silva TL, Pianca TG, Roman T, et al. Adrenergic Alpha 2A Receptor Gene and Response to Methylphenidate in Attention-Deficit/Hyperactivity Disorder Predominantly Inattentive Type. Journal of Neural Transmission. 2008; 115(2):341-345
- Stahl, S. M. Stahl’s Essential Psychopharmacology: Neuroscientific Basis and Practical Applications. Cambridge University Press. 2013.
- Cools, R. and M. D’Esposito, Inverted-U-shaped dopamine actions on human working memory and cognitive control. Biol Psychiatry, 2011. 69(12): p. e113-25.
- Cools, R., Role of dopamine in the motivational and cognitive control of behavior. Neuroscientist, 2008. 14(4): p. 381-95.
- Karczewski, KJ, Daneshjou R, Altman RB. Chapter 7: Pharmacogenomics. PLoS Comput Biol 2012;8(12):e1002817.
- Shield, A.J., et al., Human catechol O-methyltransferase genetic variation: gene resequencing and functional characterization of variant allozymes. Mol Psychiatry, 2004. 9(2): p. 151-60.
- Hain DT, Al Habbab T, Cogan ES, Johnson HL, Law RA, Lewis DJ. Review and Meta-analysis on the Impact of the ADRA2A Variant rs1800544 on Methylphenidate Outcomes in Attention-Deficit/Hyperactivity Disorder. Biol Psychiatry Glob Open Sci. 2021;2(2):106-114. Published 2021 Aug 4.