According to the American Psychiatric Association Guidelines for Major Depressive Disorders and the most recent Canadian Network for Mood and Anxiety Treatments (CANMAT), a SSRI, a SNRI, mirtazapine, or bupropion can all be considered as first-line options for the treatment of major depressive disorder (MDD).1-2
Many factors such as clinical features, medication characteristics, and patient factors influence the choice of a first-line antidepressant. SSRIs are the most commonly utilized antidepressants, however, the relative differences between first-line medications are small.3 Selecting an antidepressant involves an individualized assessment of each patient.
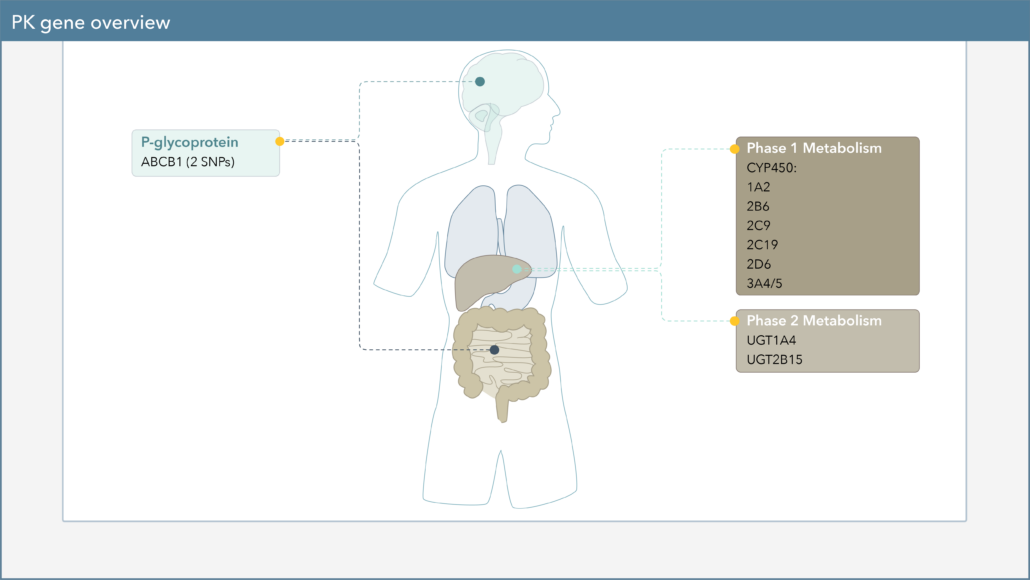
When choosing a medication to treat your patient’s major depressive disorder (MDD), there are several genetic biomarkers that may impact the response or side-effect rate of that drug. On the Genomind Pharmacogenetic Test you’ll find several pharmacokinetic (PK) genes that can impact medications commonly used for MDD.
How Pharmacokinetic Gene Variation Affects Drug Response
Pharmacokinetics, often described as “what the body does to a drug,” describes a drug’s journey throughout the body and focuses on the absorption, distribution, metabolism, and excretion of drugs.4 One of the major components of pharmacokinetics is drug metabolism, which leads to the biotransformation of drugs within the body. Genetic variation in PK genes can alter drug metabolism and consequently may influence drug efficacy and safety.
How do CYP450 Genetic Variations Affect Drug Metabolism?
Cytochrome P450 (CYP450) enzymes are major pathways in drug biotransformation and are responsible for the Phase I metabolism of many medications. In fact, 70-80% of medications are metabolized by the CYP450 system, including several psychotropic medications.5-6 Variations in these genes can lead to alterations in enzyme metabolism. Patients are typically reported as extensive/normal (EM), intermediate (IM), poor (PM), rapid (RM), or ultra-rapid metabolizers (UM).
How do CYP450 Genetic Variations Impact SSRIs?
Variations in these genes can lead to alterations in drug levels and overall drug exposure of several medications, including the commonly prescribed first-line treatment MDD, the SSRIs. Taking a more in-depth look into SSRI metabolism, Figure 1 lists the different pharmacokinetic pathways that impact drug exposure for each medication.
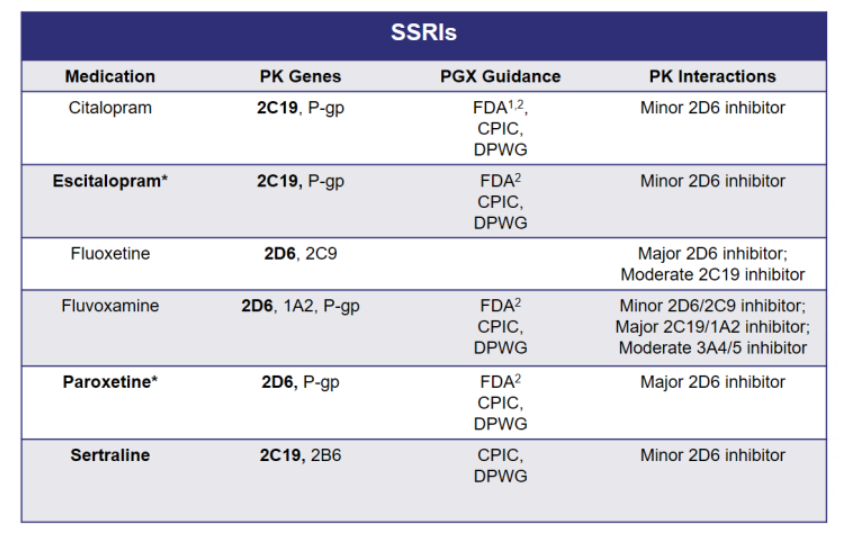
Figure 1. SSRI Metabolism
The selection of a specific SSRI can be customized to an individual patient based on the drug’s side effect profile and drug-drug interactions, but should be equally evaluated for the potential pharmacokinetic gene-drug interactions. Interindividual differences in PK parameter values and treatment outcomes with the SSRIs are associated with CYP2C19 and CYP2D6 polymorphisms.7
How do CYP2C19 Genetic Variations Affect SSRIs?
As shown in Figure 1, citalopram, escitalopram, and sertraline are extensively catalyzed by CYP2C19, thus genetic variation in CYP2C19 activity may result in altered drug exposure. Elevated concentrations of these drugs have been observed in poor metabolizers, which may increase the risk of adverse drug reactions.7
You may recall that in 2011, the FDA issued a Drug Safety Communication stating that citalopram should no longer be used at doses greater than 40mg per day. The drug label was revised to include new warnings about the potential for dose-dependent QT interval prolongation.
The FDA labeling for citalopram specifically states “…in CYP2C19 poor metabolizers, citalopram steady state Cmax and AUC was increased by 68% and 107%, respectively. Citalopram [Celexa] 20mg/day is the maximum recommended dose in CYP2C19 poor metabolizers due to the risk of QT prolongation…” 8
Citalopram is just one of many psychotropic medications that have specific dosage adjustments for CYP450 poor metabolizers in the FDA Table of Pharmacogenomic Biomarkers in Drug Labeling.
How Common are CYP2C19 Polymorphisms?
Allele frequency differs across ethnic populations. Studies have shown that roughly 13% of East Asians are CYP2C19 poor metabolizers while only 2% of Europeans are poor metabolizers. On the other hand, less than 3% of East Asians are CYP2C19 rapid metabolizers while more than 27% of Europeans are rapid metabolizers.9
While the specific guidance on citalopram dosing provided by the FDA may seem to be of limited clinical utility (i.e., one genotype, impacting one drug), users of PGx testing should also be aware there are other organizations that have reviewed the literature and published guidelines for several antidepressants outside of citalopram. For example, clinical guidance for the SSRIs based on various CYP2C19 polymorphisms is provided by the Clinical Pharmacogenetics Implementation Consortium (CPIC) and The Dutch Pharmacogenetics Working Group (DPWG).
What is CPIC?
CPIC consists of a group of researchers that specialize in pharmacogenetics from major institutions across the U.S. and several other countries . They have produced peer-reviewed guidelines and put forth standard definitions for CYP450 genotypes and associated phenotypes. As shown in Figure 2, CPIC provides dosing recommendations for citalopram, escitalopram, and sertraline based on CYP2C19 phenotype. Ultrarapid metabolizers have significantly lower exposure to these drugs (when compared to extensive metabolizers) and therefore may have increased probability of failing therapy.9
Figure 2. CPIC Dosing Recommendations for CYP2C19 and SSRIs
How do CYP2D6 Genetic Variations Affect SSRIs?
Going back to Figure 1, fluoxetine, fluvoxamine, and paroxetine are metabolized by CYP2D6. Fluoxetine metabolism is complex, as both CYP2D6 and CYP2C9 convert fluoxetine to pharmacologically active compounds. Paroxetine and fluvoxamine are extensively metabolized by CYP2D6 to compounds with little pharmacological activity towards serotonin transporters, and thus variations in CYP2D6 activity may result in altered exposure to paroxetine and fluvoxamine. As shown in Figure 3, CPIC provides dosing recommendations for paroxetine and fluvoxamine based on CYP2D6 phenotype.9
Figure 3. CPIC Dosing Recommendations for CYP2D6 and SSRIs
How do CYP450 Genetic Variations Impact Other First-line Treatments?
Variations in PK genes can lead to alterations in drug levels and overall drug exposure of several first-line options for the treatment of MDD. CYP2D6, CYP2C19, CYP3A4/5, and/or CYP1A2 are involved in the metabolism of the SNRIs. Mirtazapine, bupropion, and several atypical antidepressants are discussed in “Management of Major Depressive Disorder with a Focus on Pharmacogenetics | Part Three: The ‘Other Antidepressants.'”
In Conclusion
Variants in CYP450 enzymes can provide clinicians with greater insight into a patient’s individual drug metabolism profile. This insight may allow for more patient-specific treatment decisions and medication choices when treating MDD.
CPIC has published peer-reviewed guidelines that summarize the evidence from published literature supporting gene-drug associations. In regards to first-line antidepressants, they have guidelines on SSRIs, in which they provide dosing recommendations based on CYP2D6 and/or CYP2C19 genotype.
Pharmacogenetic testing reports from Genomind outline genetic variants in six key CYP450 genes (1A2, 2B6, 2C19, 2C9, 2D6, 3A4/3A5) and several other PK genes.
In order to help clinicians navigate the PK genes and available pharmacogenetic guidelines, Genomind provides clinicians with access to GenMedPro™, gene-drug interaction software.
GenMedPro™ serves as a comprehensive tool to help evaluate both gene-drug and drug-drug interactions, pharmacogenetic guideline recommendations, and alternative medication options, as appropriate. This robust interaction database can help clinicians assess personalized medication options and regimens for their patients. To learn more about GenMedPro™ or to request a demonstration from our PhD and PharmD experts, please utilize the Genomind Clinician Portal.
Are You Ready to Upgrade Your Practice with Genomind?
Genomind’s pharmacogenetic testing is the most advanced and comprehensive mental health pharmacogenetic test available. Get access to 24 genes related to mental health, 130+ medications, 10+ conditions, state-of-the-art tools, and 360 degrees of support! Get started today.
References
- Gelenberg AJ, Freeman MP, Markowitz JC, et al. American Psychiatric Association Practice Guideline for the Treatment of Patients With Major Depressive Disorder, Third Edition. Am J Psychiatry 2010;167(suppl):1-152.
- Kennedy SH, Lam RW, McIntyre RS, et al. Canadian Network for Mood and Anxiety Treatments (CANMAT) 2016 Clinical Guidelines for the Management of Adults with Major Depressive Disorder. Can J Psychiatry 2016;61(9):540-560.
- Cipriani A, Furukawa TA, Salanti G, et al. Comparative efficacy and acceptability of 21 antidepressant drugs for the acute treatment of adults with major depressive disorder: a systematic review and network meta-analysis. Lancet 2018;391(10128):1357-1366.
- Zhao M, Wang Y, Chen M, Wu B. Introduction to Pharmacokinetics. In: Circadian Pharmacokinetics. Wu B, Lu, D, Dong D., eds. Singapore: Springer Singapore;2020.
- Lynch T, Price A. The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am Fam Physician 2007;76(3):391-396.
- Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103-41.
- Hicks JK, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98:127–134.
- Celexa. Package insert. Forest Laboratories, Inc.; 2011
- M. Whirl-Carrillo, E.M. McDonagh, J. M. Hebert, L. Gong, K. Sangkuhl, C.F. Thorn, R.B. Altman and T.E. Klein. “Pharmacogenomics Knowledge for Personalized Medicine” Clinical Pharmacology & Therapeutics (2012) 92(4): 414-417.