Genes included on Genomind’s PGx Test
Why consider genetics when prescribing?
In many instances, the practice of psychiatry can be challenging due to the common variability in treatment response. As a result, many clinicians and patients are often left with a trial-and-error approach, even when utilizing clinical guidelines. Genomind has worked over the last decade to optimize PGx guidance for mental health care, in parallel to advancements in pharmacogenetics. These advancements and efforts can help healthcare providers get a more complete understanding of their patient to identify a more personalized treatment plan.
Pharmacogenetic testing with Genomind analyzes 26 selected genes that have been shown in numerous studies to have implications for response and/or tolerability to treatments used for various psychiatric disorders.
These 26 genes are split into the two following categories:
- Pharmacodynamic genes, which may influence the body’s biological response to a drug, such as the risk of side effects.
- Pharmacokinetic genes, which may influence how quickly or slowly the body is able to metabolize certain medications, or how much of a drug can be absorbed
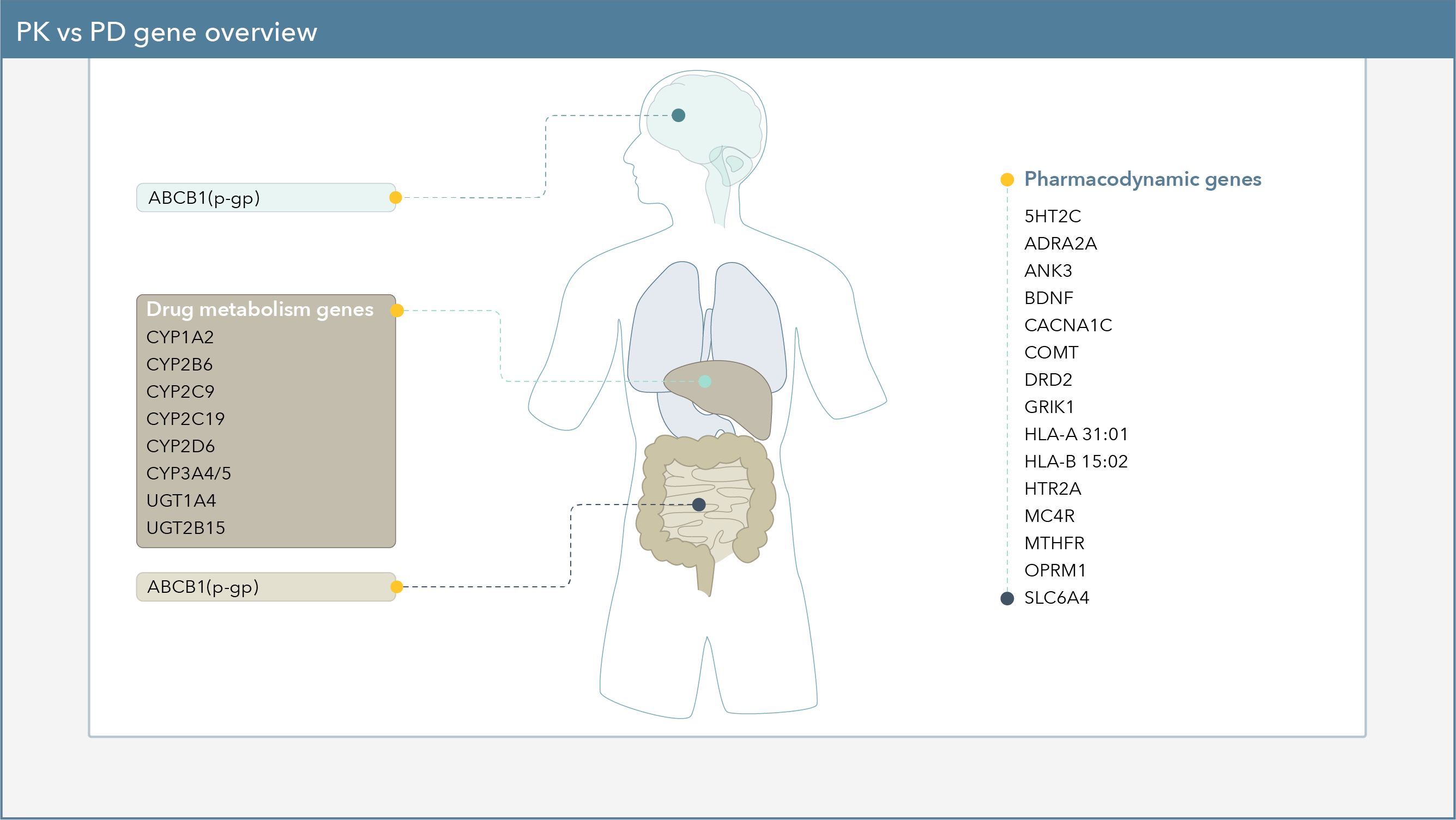
Pharmacodynamic Genes
Pharmacodynamic genes can indicate the effect that a drug has on the body and includes interactions with receptors, transporters, and neurotransmitters. Pharmacodynamic gene results provide detailed information regarding the function of the genetic variant and how the patient’s genotype may affect tolerability and/or likelihood of responding to specific therapeutic options.
Pharmacodynamic Gene by Gene Summary
SLC6A4: Serotonin Transporter
Physiological Role: The SLC6A4 gene encodes for a protein that transports serotonin from the synapse back into the presynaptic neuron. The antidepressant activity of selective serotonin reuptake inhibitors (SSRIs) is achieved primarily through inhibition of this transporter, allowing serotonin to remain in the synapse longer.
Impact of Variant: Variation in SLC6A4 influences the expression and/or production of serotonin transporters (SERTs).
Clinical Impact: Distinct genetic variations have been identified that impact the likelihood of response to and/or side effect risk with SSRIs.
DRD2: Dopamine Receptor D2
Physiological Role: The DRD2 gene encodes for a protein involved in the neurotransmission of dopamine in the brain. Most antipsychotics act primarily through modulation of the D2 receptor to reduce dopamine signaling.
Impact of Variant: A single base pair deletion (DEL) in the promoter region of the DRD2 gene has been associated with reduced DRD2 gene expression.
Clinical Impact: Carrying the DEL allele may lower the likelihood of response and/or increase weight gain with certain antipsychotic medications.
MTHFR: Methylenetetrahydrofolate Reductase
Physiological Role: The MTHFR gene encodes for an enzyme responsible for the conversion of folate (folic acid) into the biologically active form, methylfolate. Methylfolate plays a key role in the synthesis of the monoamine neurotransmitters.
Impact of Variant: C677T and A1298C variants are associated with reduced activity of the enzyme and a downstream reduction in methylfolate.
Clinical Impact: Distinct genetic variations may help identify patients with reduced capacity to produce methylfolate and may prompt consideration of l-methylfolate supplementation.
HLA-A: Major histocompatibility complex, class I, A
HLA-B: Major histocompatibility complex, class I, B
Physiological Role: HLA genes are highly polymorphic and encode for proteins that bind and present antigens to immune cells.
Impact of Variant: The HLA-B*15:02 variant has been associated with severe adverse skin reactions to certain anticonvulsants.
Clinical Impact: Carrying the HLA-B*15:02 allele impacts the likelihood of developing Stevens-Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN) with carbamazepine, oxcarbazepine, lamotrigine, phenytoin, and fosphenytoin.
BDNF: Brain-derived Neurotrophic Factor
Physiological Role: The BDNF gene encodes for a protein that is important in neuronal development and neural plasticity.
Impact of Variant: Variation in this gene is associated with altered BDNF secretion.
Clinical Impact: Carrying the Met allele can lead to a more pronounced response to exercise in improving cognition and stress response. Additionally, distinct variations in this gene are associated with an ethnicity dependent response to antidepressants. (See Ethnopsychopharmacology for more on ethnicity dependent responses to antidepressants.)
COMT: Catechol-O-Methyltransferase
Physiological Role: The COMT gene encodes for an enzyme, COMT, which breaks down dopamine in the prefrontal cortex of the brain.
Impact of Variant: Variation in this gene can impact the breakdown of dopamine in the prefrontal cortex.
Clinical Impact: Distinct genetic variations have been identified that can influence the likelihood of response to dopaminergic stimulants and certain second generation antipsychotics.
Learn more about COMT and its potential effects on stimulant response.
ADRA2A: Alpha-2A Adrenergic Receptor
Physiological Role: The ADRA2A gene encodes for a protein involved in norepinephrine signaling.
Impact of Variant: Variation within ADRA2A may influence the sensitivity of these receptors to norepinephrine.
Clinical Impact: Distinct genetic variations have been identified that can influence the likelihood of response to methylphenidate for inattentive symptoms of ADHD.
Learn more about ADRA2A and its potential impact on ADHD treatment.
MC4R: Melanocortin 4 Receptor
Physiological Role: The MC4R gene encodes for a protein involved in the control of food intake and energy expenditure.
Impact of Variant: Variation in the MC4R may cause receptor desensitization, in which the MC4R signaling is weakened.
Clinical Impact: Individuals with the A/A genotype have a higher risk of weight gain with second generation antipsychotics.
HTR2C: Serotonin Receptor 2C
Physiological Role: The HTR2C gene encodes for a protein involved in the regulation of satiety.
Impact of Variant: Variation in this gene may impact HTR2C receptor expression.
Clinical Impact: Carrying the T allele can lower the risk of weight gain with second generation antipsychotics.
ANK3: (Ankyrin-3)
Physiological Role: The ANK3 gene encodes for a protein called ankyrin G, which plays a role in sodium channel functioning and regulation of excitatory signaling.
Impact of Variant: Variation in ANK3 can lead to disturbances in sodium channel function and alterations in neuronal signaling.
Clinical Impact: Carrying the T allele may be clinically associated with conditions characterized by mood instability or lability. This may prompt consideration and evaluation of mood stabilizers if clinically indicated.
CACNA1C: Calcium Voltage-Gated Channel Subunit Alpha1 C
Physiological Role: The calcium channel gene, abbreviated CACNA1C, encodes for a protein involved in voltage-gated calcium channels. Calcium channels permit calcium ions to enter the cell, leading to excitation of neurons and brain signaling.
Impact of Variant: Variation in CACNA1C can lead to more calcium entry into the cell and alterations in neuronal signaling.
Clinical Impact: Distinct genetic variations may be clinically associated with impairment of mood or cognition. This may prompt consideration and evaluation of mood stabilizers if clinically indicated.
HTR2A: Serotonin Receptor 2A
Physiological Role: The HTR2A gene encodes for the 5-HTR2A receptors, which are downregulated by a variety of antidepressants and involved in secondary messenger cascades.
Impact of Variant: The exact mechanism by which variation in HTR2A moderates the effects of antidepressants remains undetermined.
Clinical Impact: Individuals with the A/A genotype are associated with a lower likelihood of response to antidepressants; however, additional data has supported a potential increased likelihood of response to citalopram.
OPRM1: μ-Opioid Receptor
Physiological Role: The OPRM1 gene encodes for a protein that acts as a receptor for opioids in the brain. This receptor can be affected by natural and synthetic compounds.
Impact of Variant: Variation in this gene has been linked to reduced expression of OPRM1.
Clinical Impact: Individuals with the GG genotype may have decreased sensitivity to opioids, which can result in decreased analgesia and the need for higher doses with opioids.
Learn more about how an OPRM1 variant affects individual sensitivity to opioids.
GRIK1: Glutamate Receptor Kainate 1
Physiological Role: The GRIK1 gene encodes for a protein involved in the neurotransmission of glutamate. Topiramate blocks highly selective glutamate receptors, most notably receptors within the GRIK1 subunit. GRIK1 helps to assemble these excitatory glutamate receptors, which are involved in various neurological processes.
Impact of Variant: The exact mechanism by which variation in GRIK1 moderates topiramate’s effect for alcohol use disorder remains undetermined.
Clinical Impact: Variation in this gene may help identify patients who may be more likely to benefit from topiramate for alcohol use disorder.
Pharmacokinetic Genes
Pharmacokinetic genes refer to the effect of the body on the drug, which may influence how quickly or slowly the body is able to metabolize certain medications, or how much of a drug can be absorbed.
Pharmacokinetic gene results provide detailed information regarding the function of the genetic variant and how a patient’s genotype may affect drug metabolism and drug serum levels and/or intestinal absorption and blood-brain barrier penetration of certain drugs.
Knowledge of an individual’s pharmacokinetic gene variations can help guide patient specific dosing decisions.
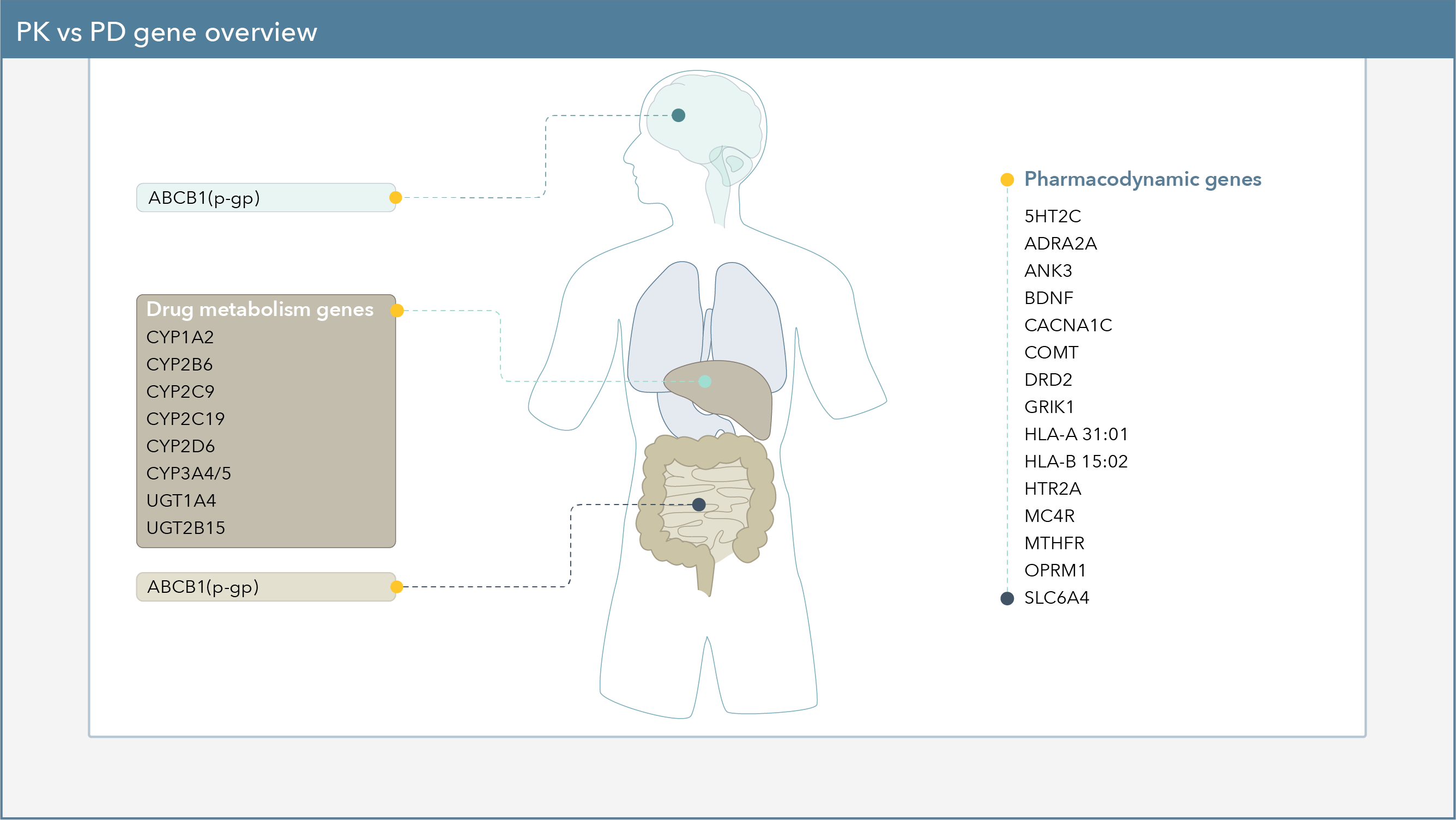
Pharmacokinetic Gene by Gene Summary
Cytochrome P450 (CYP450)
Physiological Role: The CYP450s constitute a family of hepatic enzymes that catalyze the breakdown of various substances.
Impact of Variant: Distinct genetic variations in the genes that encode these CYP450 enzymes can alter their activity.
Clinical Impact: Variation in the CYP450 genes can influence how quickly or slowly the body is able to metabolize many medications.
Uridine 5'-diphospho-glucuronosyltransferase (UGT)
Physiological Role: UGT enzymes play an important role in phase II drug metabolism.
Impact of Variant: Distinct genetic variations in the genes that encode these UGT enzymes can alter their activity.
Clinical Impact: Variation in the UGT genes can influence how quickly or slowly the body is able to metabolize certain medications.
ATP Binding Cassette Subfamily B Member 1 (ABCB1)
Physiological Role: The ABCB1 gene encodes the permeability glycoprotein, or P-glycoprotein (P-gp), an efflux pump that controls the transport of molecules across many important tissues.
Impact of Variant: Distinct genetic variations in the ABCB1 gene can influence the intestinal absorption and blood-brain barrier penetration of certain P-gp substrates.
Clinical Impact: Variation in the ABCB1 genes can influence medication response and/or tolerability of certain antidepressants, antipsychotics, and opioids.
