When choosing a medication to treat your patient’s generalized anxiety disorder (GAD), there are several genetic biomarkers that may impact drug efficacy and safety. On the Genomind pharmacogenetic test you’ll find several pharmacokinetic (PK) genes that may impact psychiatric medications commonly utilized in the treatment of GAD.
How Pharmacokinetic Gene Variation Affects Drug Response
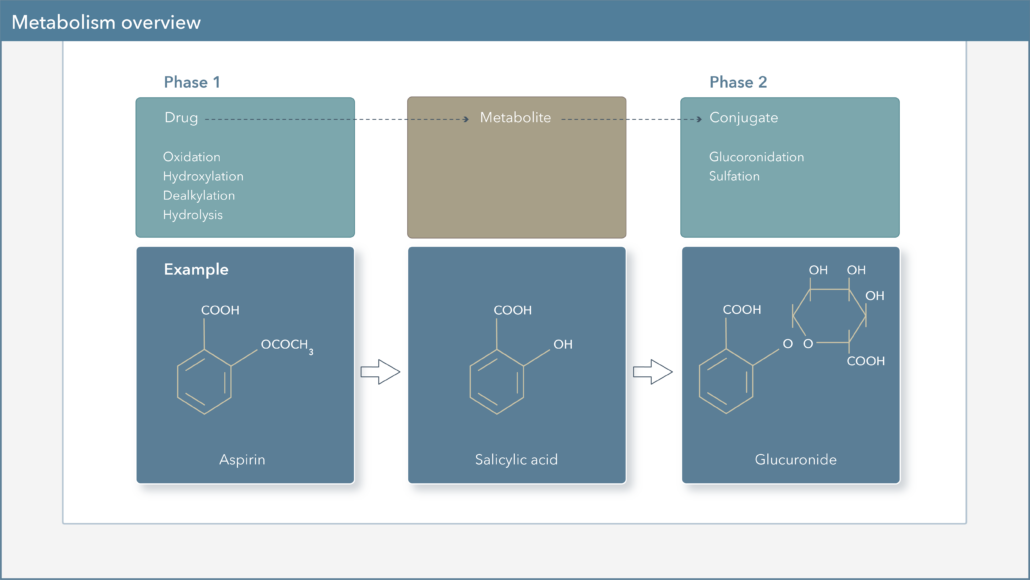
Pharmacokinetics, often described as “what the body does to a drug”, describes a drug’s journey throughout the body and focuses on the absorption, distribution, metabolism, and excretion of these compounds.1 As we discussed in Pharmacokinetic Genes in GAD Treatment: Part One, genetic variations in CYP450 genes can lead to variability in drug levels and overall drug exposure. Let’s now discuss another PK gene that can impact benzodiazepine pharmacokinetics.
How does UGT Affect Drug Metabolism?
UDP-glucuronosyltransferase (UGT) enzymes are important components of drug biotransformation, and are involved in the Phase II metabolism of many medications, which leads to the formation of more water-soluble compounds that are more easily excreted. The UGT enzyme is responsible for transferring the glucuronic acid component of UDP-glucuronic acid to a drug, in order to increase water solubility. UGT enzymes are found in several organs of the body, including the liver, skin, lungs, small intestine, and kidneys and are grouped into 2 subfamilies: UGT1 and UGT2.2,3
How do UGT Genetic Variations Affect Drug Metabolism?
Similar to genetic variation in CYP450 enzymes, mutations in the genes encoding for UGT can alter enzyme activity levels, which in turn can lead to variations in drug exposure. The Genomind pharmacogenetic test specifically reports on UGT1A4 and UGT2B15.
- UGT1A4: genetic variation may result in ultra-rapid metabolizer (UM) status and decreased serum levels of lamotrigine, asenapine, and trifluoperazine.
- UGT2B15: genetic variation may result in intermediate metabolizer (IM) status and subsequent increased serum levels of some benzodiazepines.
How do UGT Genetic Variations Affect Benzodiazepines?
As shown in Table 1 below, many of the benzodiazepines commonly used in anxiety disorders are metabolized by UGT2B15.
|
Table 1: Benzodiazepine Metabolism |
||
|
Benzodiazepine |
Major Active Metabolite |
Metabolism |
|
Alprazolam |
N/A |
3A4/5 |
|
Chlordiazepoxide |
Yes |
3A4/5, UGT2B15 |
|
Clonazepam |
N/A |
3A4/5 |
|
Clorazepate |
Yes |
UGT2B15 |
|
Diazepam |
Yes |
2C19, 3A4/5, UGT2B15 |
|
Lorazepam |
N/A |
UGT2B15 |
|
Oxazepam |
N/A |
UGT2B15 |
Both in vitro and in vivo studies observed the impact of the UGT2B15 polymorphism on oxazepam and lorazepam drug exposure. As an example, in a study of 24 healthy subjects, individuals with the decreased activity UGT2B15*2/*2 genotype were found to have a 0.58-fold lower clearance of intravenous lorazepam compared to those with the normal activity *1/*1 genotype.4-7
Oxazepam is an active metabolite of several other benzodiazepines (e.g., chlordiazepoxide, clorazepate, diazepam, and temazepam), thus drug exposure of these compounds may be affected by UGT2B15 polymorphisms as well.
How do CYP Genetic Variations Affect Benzodiazepines?
As shown in Table 1, many of the benzodiazepines commonly used in anxiety disorders are metabolized by CYP3A4/5. Diazepam metabolism involves CYP2C19, CYP3A4/5, and UGT2B15. Diazepam is listed in the FDA Table of Pharmacogenomic Biomarkers in Drug Labeling as well as the FDA Table of Pharmacogenetic Associations as systemic concentrations may be affected in CYP2C19 poor metabolizers.8
How do PK Drug Interactions Affect Benzodiazepines?
Various medications can act as inhibitors or inducers of CYP450 enzymes, leading to alterations in the activity of these enzymes and their ability to metabolize other medications.9 This, in turn, can lead to increased or decreased concentrations of other medications, which may be associated with toxicities or changes in efficacy, respectively.
Many prescribers find it advantageous that benzodiazepines such as lorazepam and oxazepam are not metabolized by the CYP450 enzymes. While they are less likely to be influenced by CYP interactions, it is important to recognize that drug-drug interactions can also involve UGT.
- UGT Inhibitors: cannabidiol, carvedilol, divalproex, fluconazole, ketoconazole, levothyroxine, quinidine, and valproate.
- UGT Inducers: carbamazepine, dexamethasone, fosphenytoin, and phenytoin.
In order to help clinicians navigate the CYP450 and UGT enzymes (as well as other pharmacokinetic considerations), Genomind provides clinicians with access to GenMedPro™ gene-drug interaction software.
In Conclusion
Knowledge of an individual’s variants in CYP450 and UGT enzymes can provide clinicians with greater insight into a patient’s individual drug metabolism profile. This insight may allow for more patient-specific treatment decisions and medication choices.
GenMedPro™ serves as an outstanding tool to help evaluate both gene-drug and drug-drug interactions, as well as alternative medication options, as appropriate. This robust interaction database can facilitate clinician decision-making when choosing medication options and regimens for their patients. To learn more about GenMedPro™ or to request a demonstration from our PhD and PharmD experts, please utilize Genomind’s Clinician Portal.
Are You Ready to Upgrade Your Practice with Genomind?
Genomind’s pharmacogenetic testing is the most advanced and comprehensive mental health pharmacogenetic test available. Get access to 24 genes related to mental health, 130+ medications, 10+ conditions, state-of-the-art tools, and 360 degrees of support. Register to order today.
References
- Zhao M, Wang Y, Chen M, Wu B. Introduction to Pharmacokinetics. In: Circadian Pharmacokinetics. Wu B, Lu, D, Dong D., eds. Singapore: Springer Singapore;2020.
- de Leon J. Gulcuronidation enzymes, genes and psychiatry. Int J Neuropsychopharmacol 2003;6(1):57-72.
- King, C.D., et al., UDP-glucuronosyltransferases. Curr Drug Metab, 2000. 1(2): p. 143-61.
- Chung, J.Y., et al., Effect of the UGT2B15 genotype on the pharmacokinetics, pharmacodynamics, and drug interactions of intravenous lorazepam in healthy volunteers. Clin Pharmacol Ther, 2005. 77(6): p.486-94.
- Stingl, J.C., et al., Relevance of UDP-glucuronosyltransferase polymorphisms for drug dosing: A quantitative systematic review. Pharmacol Ther, 2014. 141(1): p. 92-116.
- He, X., et al., Evidence for oxazepam as an in vivo probe of UGT2B15: oxazepam clearance is reduced by UGT2B15 D85Y polymorphism but unaffected by UGT2B17 deletion. Br J Clin Pharmacol, 2009. 68(5): p. 721-30.
- Court, M.H., et al., UDP-glucuronosyltransferase (UGT) 2B15 pharmacogenetics: UGT2B15 D85Y genotype and gender are major determinants of oxazepam glucuronidation by human liver. J Pharmacol Exp Ther, 2004. 310(2): p. 656-65.
- M. Whirl-Carrillo, E.M. McDonagh, J. M. Hebert, L. Gong, K. Sangkuhl, C.F. Thorn, R.B. Altman and T.E. Klein. “Pharmacogenomics Knowledge for Personalized Medicine” Clinical Pharmacology & Therapeutics (2012) 92(4): 414-417.
- Phang-Lyn S, Llerena VA. Biochemistry, Biotransformation. [Updated 2020 Sep 2]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2020 Jan.